Visualization:
Code for general neural data plotting (raster plots, multi-channel field potential plots, psth, etc.)
API
Base
- aopy.visualization.base.advance_plot_color(ax, n)[source]
Utility to skip colors for the given axis.
- Parameters:
ax (pyplot.Axes) – specify which axis to advance the color
n (int) – how many colors to skip in the cycle
Examples
Using advance_plot_color to skip the first color in the cycle.
plt.subplots() aopy.visualization.advance_plot_color(plt.gca(), 1) plt.plot(np.arange(10), np.arange(10))

- aopy.visualization.base.annotate_spatial_map(elec_pos, text, color, fontsize=6, ax=None, **kwargs)[source]
Simple wrapper around plt.annotate() to add text annotation to a 2d position.
- Parameters:
elec_pos ((x,y) tuple) – position where text should be placed on 2d plot
text (str) – annotation text
color (plt.Color) – the color to make the text
fontsize (int, optional) – the fontsize to make the text. Defaults to 6.
ax (pyplot.Axes, optional) – axis on which to plot. Defaults to None.
kwargs (dict) – additional keyword arguments to pass to plt.annotate()
- Returns:
annotation object
- Return type:
plt.Annotation
- aopy.visualization.base.annotate_spatial_map_channels(acq_idx=None, acq_ch=None, drive_type='ECoG244', theta=0, color='k', fontsize=6, ax=None, **kwargs)[source]
Given acq_idx (indices) or acq_ch (channel numbers), prints either indices or channel numbers on top of a spatial map.
- Parameters:
acq_idx ((nacq,) array or list, optional) – If provided, specifies the acquisition indices to be annotated. If neither acq_idx nor acq_ch is provided, all channel numbers will be annotated by default.
acq_ch ((nacq,) array or list, optional) – If provided, specifies the acquisition channel numbers to be annotated. If neither acq_idx nor acq_ch is provided, all channel numbers will be annotated by default.
drive_type (str, optional) – Drive type of the channels to plot. See
aopy.data.base.load_chmap().color (str, optional) – color to display the channels. Default ‘k’.
fontsize (int, optional) – the fontsize to make the text. Defaults to 6.
print_zero_index (bool, optional) – if True (the default), prints channel numbers indexed by 0. Otherwise prints directly from the channel map (which should use 1-indexing).
ax (pyplot.Axes, optional) – axis on which to plot. Defaults to None.
kwargs (dict) – additional keyword arguments to pass to plt.annotate()
Example
aopy.visualization.plot_ECoG244_data_map(np.zeros(256,), cmap='Greys') aopy.visualization.annotate_spatial_map_channels(drive_type='ECoG244', color='k') aopy.visualization.annotate_spatial_map_channels(drive_type='Opto32', color='b') plt.axis('off')

Note
The acq_ch returned from func::aopy.data.load_chmap are generally 1-indexed lists of acquisition channels connected to electrodes. In python, however, the acquisition indices start at 0, so we give the option to select channels based on either an index (acq_idx) or a channel number (acq_ch).
- aopy.visualization.base.calc_data_map(data, x_pos, y_pos, grid_size, interp_method='nearest', threshold_dist=None)[source]
Turns scatter data into grid data by interpolating up to a given threshold distance.
Example
Make a plot of a 10 x 10 grid of increasing values with some missing data.
data = np.linspace(-1, 1, 100) x_pos, y_pos = np.meshgrid(np.arange(0.5,10.5),np.arange(0.5, 10.5)) missing = [0, 5, 25] data_missing = np.delete(data, missing) x_missing = np.reshape(np.delete(x_pos, missing),-1) y_missing = np.reshape(np.delete(y_pos, missing),-1) interp_map, xy = calc_data_map(data_missing, x_missing, y_missing, [10, 10], threshold_dist=1.5) plot_spatial_map(interp_map, xy[0], xy[1])

- Parameters:
data (nch) – list of values
x_pos (nch) – list of x positions
y_pos (nch) – list of y positions
grid_size (tuple) – number of points along each axis
interp_method (str) – method used for interpolation
threshold_dist (float) – distance to neighbors before disregarding a point on the image
- Returns:
- tuple containing:
- data_map (grid_size array, e.g. (16,16)): map of the data on the given gridxy (grid_size array, e.g. (16,16)): new grid positions to use with this map
- Return type:
tuple
- aopy.visualization.base.color_trajectories(trajectories, labels, colors, ax=None, **kwargs)[source]
Draws the given trajectories but with the color of each trajectory corresponding to its given label. Works for 2D and 3D axes
Example
trajectories =[ np.array([ [0, 0, 0], [1, 1, 0], [2, 2, 0], [3, 3, 0], [4, 2, 0] ]), np.array([ [-1, 1, 0], [-2, 2, 0], [-3, 3, 0], [-3, 4, 0] ]) ] labels = [0, 0, 1, 0] colors = ['r', 'b'] color_trajectories(trajectories, labels, colors) .. image:: _images/color_trajectories_simple.png labels_list = [[0, 0, 1, 1, 1], [0, 0, 1, 1], [1, 1, 0, 0]] fig = plt.figure() color_trajectories(trajectories, labels_list, colors) .. image:: _images/color_trajectories_segmented.png
- Parameters:
trajectories (ntrials) – list of (n, 2) or (n, 3) trajectories where n can vary across each trajectory
labels (ntrials) – integer array of labels for each trajectory. Basically an index for each trajectory
colors (ncolors) – list of colors. A list of arrays containing the label for each corresponding trajectory, or a list of lists where each sublist corresponds to the label for each timepoint in the corresponding trajectory.
ax (plt.Axis, optional) – axis to plot the targets on
**kwargs (dict) – other arguments for plot_trajectories(), e.g. bounds
- aopy.visualization.base.combine_channel_figures(figuredir, nch=256, figsize=(6, 5), dpi=150)[source]
Combines all channel figures in directory generated from plot_channel_summary
- Parameters:
figuredir (str) – path to directory of channel profile images
nch (int, optional) – number of channels from data array. Determines combined image layout. Defaults to 256.
figsize (tuple, optional) – (width, height) to pass to pyplot. Default (6, 5)
dpi (int, optional) – resolution to pass to pyplot. Default 150
- aopy.visualization.base.get_color_gradient_RGB(npts, end_color, start_color=[1, 1, 1])[source]
This function outputs an ordered list of RGB colors that are linearly spaced between white and the input color. See also sns.color_palette for a gradient of RGB values within a Seaborn color palette.
Examples
npts = 200 x = np.linspace(0, 2*np.pi, npts) y = np.sin(x) fig, ax = plt.subplots() ax.scatter(x, y, c=get_color_gradient(npts, 'g', [1,0,0]))

- Parameters:
npts (int) – How many different colors are part of the gradient
end_color (str or list) – Color that ends the gradient. Can be any matplotlib color or specific RGB values.
start_color (str or list) – Color that ends the gradient. Can be any matplotlib color or specific RGB values. Defaults to white.
- Returns:
An array with linearly spaced colors from the start to end
- Return type:
(npts, 3)
- aopy.visualization.base.get_data_map(data, x_pos, y_pos)[source]
Organizes data according to the given x and y positions
- Parameters:
data (nch) – list of values
x_pos (nch) – list of x positions
y_pos (nch) – list of y positions
- Returns:
map of the data on the grid defined by x_pos and y_pos
- Return type:
(m,n array)
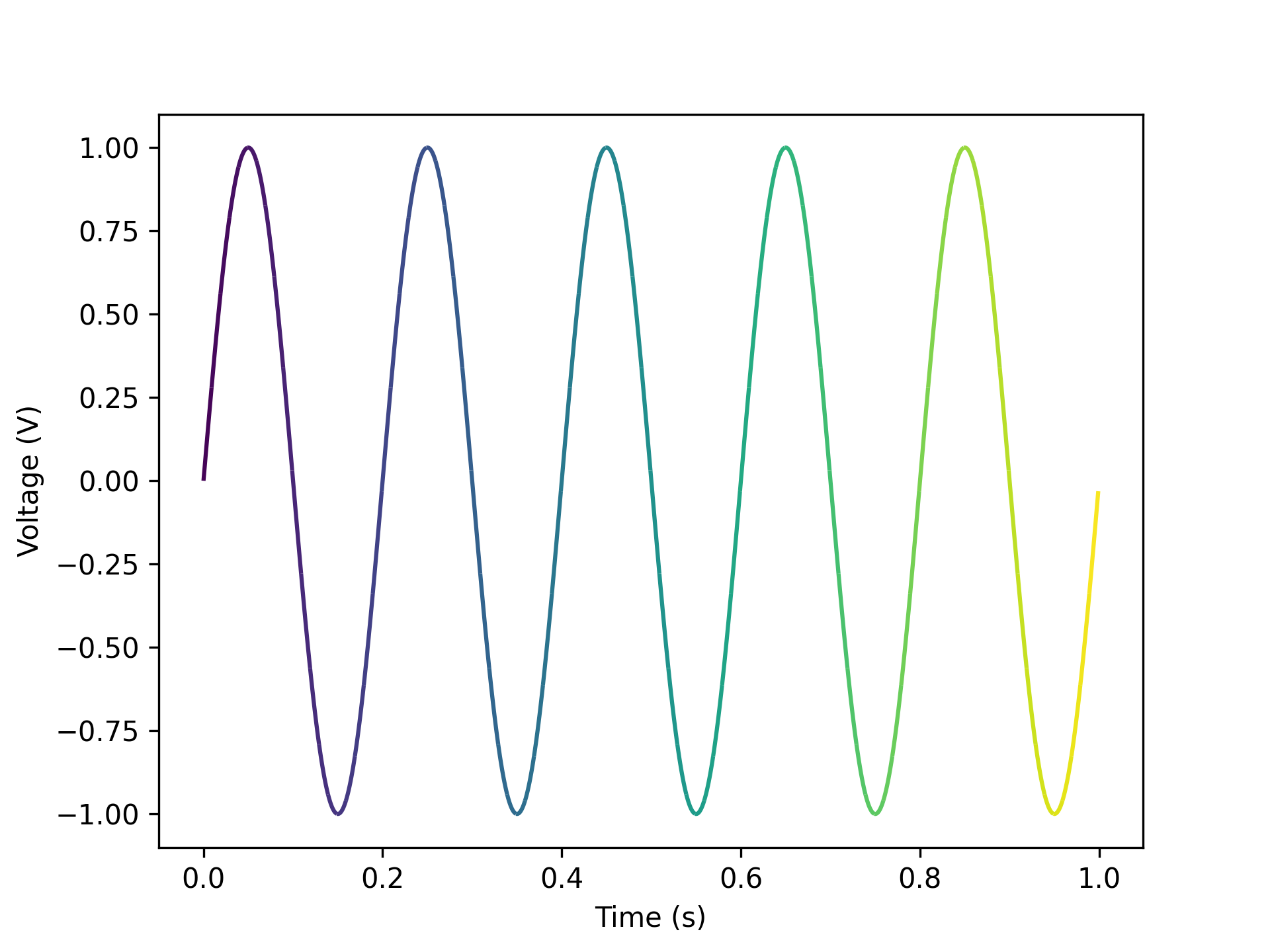
- aopy.visualization.base.gradient_timeseries(data, samplerate, n_colors=100, color_palette='viridis', ax=None, **kwargs)[source]
Draw gradient lines of timeseries data. Default units are seconds and volts.
- Parameters:
data (nt, nch) – timeseries to plot, can be 1d or 2d.
samplerate (float) – sampling rate of the data
n_colors (int, optional) – number of colors in the gradient. Default 100.
color_palette (str, optional) – colormap to use for the gradient. Default ‘viridis’.
ax (plt.Axis, optional) – axis to plot the targets on
kwargs (dict) – keyword arguments to pass to the LineCollection function (similar to plt.plot)
- Raises:
ValueError – if the data has more than 2 dimensions
Example
data = np.reshape(np.sin(np.pi*np.arange(1000)/100), (1000)) samplerate = 1000 gradient_timeseries(data, samplerate)
- aopy.visualization.base.gradient_trajectories(trajectories, n_colors=100, color_palette='viridis', bounds=None, ax=None, **kwargs)[source]
Draw trajectories with a gradient of color from start to end of each trajectory. Works in 2D and 3D.
- Note: this function applies the gradient evenly across the timepoints of the trajectory.
It might be useful to use the sampling rate of the data instead of n_colors, so that the time axis is consistent across sampling rates.
- Parameters:
trajectories (ntrials) – list of 2D or 3D trajectories, in x, y[, z] coordinates
n_colors (int, optional) – number of colors in the gradient. Default 100.
color_palette (str, optional) – colormap to use for the gradient. Default ‘viridis’.
bounds (tuple, optional) – 6-element tuple describing (-x, x, -y, y, -z, z) axes bounds
ax (plt.Axis, optional) – axis to plot the targets on
kwargs (dict) – keyword arguments to pass to the LineCollection function (similar to plt.plot)
Example
Cursor trajectories in 2D .. code-block:: python
subject = ‘beignet’ te_id = 5974 date = ‘2022-07-01’ preproc_dir = data_dir traj, _ = aopy.data.get_kinematic_segments(preproc_dir, subject, te_id, date, [32], [81, 82, 83, 239]) gradient_trajectories(traj[:3])

Hand trajectories in 3D .. code-block:: python
traj, _ = aopy.data.get_kinematic_segments(preproc_dir, subject, te_id, date, [32], [81, 82, 83, 239], datatype=’hand’) plt.figure() ax = plt.axes(projection=’3d’) gradient_trajectories(traj[:3], bounds=[-10,0,60,70,20,40], ax=ax)

Note
Automatic bounds aren’t set in 3D plots. The best alternative is to first plot in 2D, then use those bounds to manually set the first 2 axes bounds for the 3D plot.
- aopy.visualization.base.place_Opto32_subplots(fig_size=5, subplot_size=0.75, offset=(0.0, -0.25), **kwargs)[source]
Wrapper around place_subplots() for the Opto32 stimulation sites.
- Parameters:
fig_size (float) – width and height (in inches) of the figure
subplot_size (float) – width and height (in inches) of each subplot
offset (tuple) – x and y offset (in inches) from the bottom left corner of the figure
kwargs (dict, optional) – other keyword arguments to pass to fig.add_axes
- Returns:
tuple containing: | fig (pyplot.Figure): figure where the subplots were placed | ax (list): pyplot.Axes handles for each stimulation site
- Return type:
tuple
Examples

- aopy.visualization.base.place_subplots(fig, positions, width, height, **kwargs)[source]
Plotting utility to create subplots in arbitrary positions on a figure. Positions are in inches from the bottom left corner of the figure.
- Parameters:
fig (pyplot.Figure) – figure to place the subplots on
positions (npos, 2) – list of (x, y) coordinates (in inches) where to center the subplots
width (float) – width (in inches) of each subplot
height (float) – height (in inches) of each subplot
kwargs (dict, optional) – other keyword arguments to pass to fig.add_axes
- Returns:
pyplot.Axes handles for each position
- Return type:
list
Examples
fig = plt.figure(figsize=(4,6)) positions = [[1, 2], [3, 4]] width = 1 height = 1 ax = place_subplots(fig, positions, width, height) ax[0].annotate('1', (0.5,0.5), fontsize=40) ax[1].annotate('2', (0.5,0.5), fontsize=40)

fig = plt.figure(figsize=(4,6)) positions = [[1, 1.5], [3, 4.5]] width = 2 height = 3 ax = place_subplots(fig, positions, width, height) ax[0].annotate('1', (0.5,0.5), fontsize=40) ax[1].annotate('2', (0.5,0.5), fontsize=40)

- aopy.visualization.base.plot_ECoG244_data_map(data, bad_elec=[], interp=True, cmap='bwr', theta=0, ax=None, **kwargs)[source]
Plot a spatial map of data from an ECoG244 electrode array from the Viventi lab.
- Parameters:
data ((256,) array) – values from the ECoG array to plot in 2D
bad_elec (list, optional) – channels to remove from the plot. Defaults to [].
interp (bool, optional) – flag to include 2D interpolation of the result. Defaults to True.
cmap (str, optional) – matplotlib colormap to use in image. Defaults to ‘bwr’.
theta (float) – rotation (in degrees) to apply to positions. rotations are applied clockwise, e.g., theta = 90 rotates the map clockwise by 90 degrees, -90 rotates the map anti-clockwise by 90 degrees. Default 0.
ax (pyplot.Axes, optional) – axis on which to plot. Defaults to None.
kwargs (dict) – dictionary of additional keyword argument pairs to send to calc_data_map and plot_spatial_map.
- Returns:
image returned by pyplot.imshow. Use to add colorbar, etc.
- Return type:
pyplot.Image
Examples
data = np.linspace(-1, 1, 256) missing = [0, 5, 25] plt.figure() plot_ECoG244_data_map(data, bad_elec=missing, interp=False, cmap='bwr', ax=None) # Here the missing electrodes (in addition to the ones # undefined by the channel mapping) should be visible in the map. plt.figure() plot_ECoG244_data_map(data, bad_elec=missing, interp=False, cmap='bwr', ax=None, nan_color=None) # Now we make the missing electrodes transparent plt.figure() plot_ECoG244_data_map(data, bad_elec=missing, interp=True, cmap='bwr', ax=None) # Missing electrodes should be filled in with linear interp.
- aopy.visualization.base.plot_boxplots(data, plt_xaxis, trendline=True, facecolor='gray', linecolor='k', box_width=0.5, ax=None)[source]
This function creates a boxplot for each column of input data. If the input data has NaNs, they are ignored.
- Parameters:
data (ncol list or (m, ncol) array) – Data to plot. A different boxplot is created for each entry of the list.
plt_xaxis (ncol) – X-axis locations or labels to plot the boxplot of each column
trendline (bool) – If a line should be used to connect boxplots
facecolor (color) – Color of the box faces. Can be any input that pyplot interprets as a color.
linecolor (color) – Color of the connecting lines.
ax (axes handle) – Axes to plot
Examples
Using a rectangular array and numeric x-axis points.
data = np.random.normal(0, 2, size=(20, 5)) xaxis_pts = np.array([2,3,4,4.75,5.5]) fig, ax = plt.subplots(1,1) plot_boxplots(data, xaxis_pts, ax=ax)

Using a list of nonrectangular arrays with categorical x-axis points.
data = [np.random.normal(0, 2, size=(10)), np.random.normal(0, 1, size=(20))] xaxis_pts = ['foo', 'bar'] fig, ax = plt.subplots(1,1) plot_boxplots(data, xaxis_pts, ax=ax)

- aopy.visualization.base.plot_channel_summary(chdata, samplerate, nperseg=None, noverlap=None, trange=None, title=None, figsize=(6, 5), dpi=150, frange=(0, 80), cmap_lim=(0, 40))[source]
Plot time domain trace, spectrogram and normalized (z-scored) spectrogram. Computes spectrogram.
--------------- | time series | |-------------| | spectrogram | |-------------| | norm sgram | ---------------
- Parameters:
chdata (nt,1) – neural recording data from a given channel (lfp, ecog, broadband)
samplerate (int) – data sampling rate
nperseg (int) – length of each spectrogram window (in samples)
noverlap (int) – number of samples shared between neighboring spectrogram windows (in samples)
trange (tuple, optional) – (min, max) time range to display. Default show the entire time series
title (str, optional) – print a title above the timeseries data. Default None
figsize (tuple, optional) – (width, height) to pass to pyplot. Default (6, 5)
dpi (int, optional) – resolution to pass to pyplot. Default 150
frange (tuple, optional) – range of frequencies to display in spectrogram. Default (0, 80)
cmap_lim (tuple, optional) – clim to display in the spectrogram. Default (0, 40)
- Outputs:
fig (Figure): Figure object
- aopy.visualization.base.plot_circles(circle_positions, circle_radius, circle_color='b', bounds=None, alpha=0.5, ax=None, unique_only=True)[source]
Add circles to an axis. Works for 2D and 3D axes
- Parameters:
circle_positions (ntarg, 3) – array of target (x, y, z) locations
circle_radius (float) – radius of each target
circle_color (str) – color to draw circle - default is blue
bounds (tuple, optional) – 6-element tuple describing (-x, x, -y, y, -z, z) cursor bounds
origin (tuple, optional) – (x, y, z) position of the origin
ax (plt.Axis, optional) – axis to plot the targets on
unique_only (bool, optional) – If True, function will only plot targets with unique positions (default: True)
- aopy.visualization.base.plot_corr_across_entries(preproc_dir, subjects, ids, dates, band=(70, 200), taper_len=0.1, num_seconds=60, cmap='viridis', ax=None, remove_bad_ch=True, **bad_ch_kwargs)[source]
Plot the correlation vs electrode distance for each entry in the given list of subjects, ids, and dates.
- Parameters:
preproc_dir (str) – path to the preprocessed data directory
subjects (list) – list of subject names
ids (list) – list of te_ids
dates (list) – list of dates
band (tuple, optional) – frequency band to filter the data. Default (70, 200)
taper_len (float, optional) – length of taper to use in the filter. Default 0.1
num_seconds (int, optional) – number of seconds to use in the correlation calculation. Default 60
cmap (str, optional) – colormap to use for plotting. Default ‘viridis’
ax (pyplot.Axes, optional) – axis on which to plot. Default current axis
remove_bad_ch (bool, optional) – whether to remove bad channels from the data. Default True
bad_ch_kwargs (dict, optional) – keyword arguments to pass to :func:`a
Example
Plotting the correlation vs electrode distance for a few entries in the preprocessed data directory.

- aopy.visualization.base.plot_corr_over_elec_distance(elec_data, elec_pos, ax=None, **kwargs)[source]
Makes a plot of correlation vs electrode distance for the given data.
- Parameters:
elec_data (nt, nelec) – electrode data with nch corresponding to elec_pos
elec_pos (nelec, 2) – x, y position of each electrode
ax (pyplot.Axes, optional) – axis on which to plot
kwargs (dict, optional) – other arguments to supply to
aopy.analysis.calc_corr_over_elec_distance()
Example
Using the multichannel test data generator in utils, we get a phase-shifted sine wave in each channel. Assigning each channel i to an electrode with position (i, 0), the correlation across distances looks like this:
duration = 0.5 samplerate = 1000 n_channels = 30 frequency = 100 amplitude = 0.5 acq_data = aopy.utils.generate_multichannel_test_signal(duration, samplerate, n_channels, frequency, amplitude) acq_ch = (np.arange(n_channels)+1).astype(int) elec_pos = np.stack((range(n_channels), np.zeros((n_channels,))), axis=-1) plt.figure() plot_corr_over_elec_distance(acq_data, acq_ch, elec_pos)

- Updated:
2024-03-13 (LRS): Changed input from acq_data and acq_ch to elec_data.
- aopy.visualization.base.plot_events_time(events, event_timestamps, labels, ax=None, colors=['tab:blue', 'tab:orange', 'tab:green'])[source]
This function plots multiple different events on the same plot. The first event (item in the list) will be displayed on the bottom of the plot.

- Parameters:
events (list (nevents) of 1D arrays (ntime)) – List of Logical arrays that denote when an event(for example, a reward) occurred during an experimental session. Each item in the list corresponds to a different event to plot.
event_timestamps (list (nevents) of 1D arrays ntime) – List of 1D arrays of timestamps corresponding to the events list.
labels (list (nevents) of str) – Event names for each list item.
ax (axes handle) – Axes to plot
colors (list of str) – Color to use for each list item
- aopy.visualization.base.plot_freq_domain_amplitude(data, samplerate, ax=None, rms=False)[source]
Plots a amplitude spectrum of each channel on the given axis. Just need to input time series data and this will calculate and plot the amplitude spectrum.
Example
Plot 50 and 100 Hz sine wave amplitude spectrum.
data = np.sin(np.pi*np.arange(1000)/10) + np.sin(2*np.pi*np.arange(1000)/10) samplerate = 1000 plot_freq_domain_amplitude(data, samplerate) # Expect 100 and 50 Hz peaks at 1 V each

- Parameters:
data (nt, nch) – timeseries data in volts, can also be a single channel vector
samplerate (float) – sampling rate of the data
ax (pyplot axis, optional) – where to plot
rms (bool, optional) – compute root-mean square amplitude instead of peak amplitude
- aopy.visualization.base.plot_image_by_time(time, image_values, ylabel='trial', cmap='bwr', ax=None)[source]
Makes an nt x ntrial image colored by the timeseries values.
Example
time = np.array([-2, -1, 0, 1, 2, 3]) data = np.array([[0, 0, 1, 1, 0, 0], [0, 0, 0, 1, 1, 0]]).T plot_image_by_time(time, data) filename = 'image_by_time.png'

- Parameters:
time (nt) – time vector to plot along the x axis
image_values (nt, [nch or ntr]) – time-by-trial or time-by-channel data
ylabel (str, optional) – description of the second axis of image_values. Defaults to ‘trial’.
cmap (str, optional) – colormap with which to display the image. Defaults to ‘bwr’.
ax (pyplot.Axes, optional) – Axes object on which to plot. Defaults to None.
- Returns:
the image object returned by pyplot
- Return type:
pyplot.AxesImage
- aopy.visualization.base.plot_laser_sensor_alignment(sensor_volts, samplerate, stim_times, ax=None)[source]
Plot laser sensor data aligned to the stimulus times. Useful to debug laser timing issues to make sure the laser is actually on when you think it is.
- Parameters:
sensor_volts ((nstim,) float array) – laser sensor data
samplerate (float) – sampling rate of the sensor data
stim_times ((nstim,) array) – times at which the laser was turned on
ax (pyplot.Axes, optional) – axes on which to plot. Default current axis.
kwargs (dict, optional) – other keyword arguments to pass to pyplot
- Returns:
image object returned from pyplot.pcolormesh. Useful for adding colorbars, etc.
- Return type:
pyplot.Image
Examples

- aopy.visualization.base.plot_mean_fr_per_target_direction(means_d, neuron_id, ax, color, this_alpha, this_label)[source]
generate a plot of mean firing rate per target direction
- aopy.visualization.base.plot_raster(data, cue_bin=None, ax=None)[source]
Create a raster plot for binary input data and show the relative timing of an event with a vertical red line

- Parameters:
data (ntime, ncolumns) – 2D array of data. Typically a time series of spiking events across channels or trials (not spike count- must contain only 0 or 1).
cue_bin (float) – time bin at which an event occurs. Leave as ‘None’ to only plot data. For example: Use this to indicate ‘Go Cue’ or ‘Leave center’ timing.
ax (plt.Axis) – axis to plot raster plot
- Returns:
raster plot plotted in appropriate axis
- Return type:
None
- aopy.visualization.base.plot_sessions_by_date(trials, dates, *columns, method='sum', labels=None, ax=None)[source]
Plot session data organized by date and aggregated such that if there are multiple rows on a given date they are combined into a single value using the given method. If the method is ‘mean’ then the values will be averaged for each day, for example for size of cursor. The average is weighted by the number of trials in that session. If the method is ‘sum’ then the values will be added together on each day, for example for number of trials.
Example
Plotting success rate averaged across days.
from datetime import date, timedelta date = [date.today() - timedelta(days=1), date.today() - timedelta(days=1), date.today()] success = [70, 65, 65] trials = [10, 20, 10] fig, ax = plt.subplots(1,1) plot_sessions_by_date(trials, dates, success, method='mean', labels=['success rate'], ax=ax) ax.set_ylabel('success (%)')

- Parameters:
trials (nsessions) –
dates (nsessions) –
*columns (nsessions) – dataframe columns or numpy arrays to plot
method (str, optional) – how to combine data within a single date. Can be ‘sum’ or ‘mean’.
labels (list, optional) – string label for each column to go into the legend
ax (pyplot.Axes, optional) – axis on which to plot
- aopy.visualization.base.plot_sessions_by_trial(trials, *columns, dates=None, smoothing_window=None, labels=None, ax=None, **kwargs)[source]
Plot session data by absolute number of trials completed. Optionally split up the sessions by date and apply smoothing to each day’s data.
Example
Plotting success rate over three sessions.
success = [70, 65, 60] trials = [10, 20, 10] fig, ax = plt.subplots(1,1) plot_sessions_by_trial(trials, success, labels=['success rate'], ax=ax) ax.set_ylabel('success (%)')

- Parameters:
trials (nsessions) – number of trials in each session
*columns (nsessions) – dataframe columns or numpy arrays to plot
dates (nsessions, optional) – dataframe columns or numpy arrays of the date of each session
smoothing_window (int, optional) – number of trials to smooth. Default no smoothing.
labels (list, optional) – string label for each column to go into the legend
ax (pyplot.Axes, optional) – axis on which to plot
- aopy.visualization.base.plot_spatial_map(data_map, x, y, alpha_map=None, ax=None, cmap='bwr', nan_color='black', clim=None)[source]
Wrapper around plt.imshow for spatial data
- Parameters:
data_map ((2,n) array) – map of x,y data
x (list) – list of x positions
y (list) – list of y positions
alpha_map ((2,n) array) – map of alpha values (optional, default alpha=1 everywhere). If the alpha values are outside of the range (0,1) they will be scaled automatically.
ax (int, optional) – axis on which to plot, default gca
cmap (str, optional) – matplotlib colormap to use in image. default ‘bwr’
nan_color (str, optional) – color to plot nan values, or None to leave them invisible. default ‘black’
clim ((2,) tuple) – (min, max) to set the c axis limits. default None, show the whole range
- Returns:
image object which you can use to add colorbar, etc.
- Return type:
mappable
Examples
Make a plot of a 10 x 10 grid of increasing values with some missing data.
data = np.linspace(-1, 1, 100) x_pos, y_pos = np.meshgrid(np.arange(0.5,10.5),np.arange(0.5, 10.5)) missing = [0, 5, 25] data_missing = np.delete(data, missing) x_missing = np.reshape(np.delete(x_pos, missing),-1) y_missing = np.reshape(np.delete(y_pos, missing),-1) data_map = get_data_map(data_missing, x_missing, y_missing) plot_spatial_map(data_map, x_missing, y_missing)

Make the same image but include a transparency layer
data = np.linspace(-1, 1, 100) x_pos, y_pos = np.meshgrid(np.arange(0.5,10.5),np.arange(0.5, 10.5)) missing = [0, 5, 25] data_missing = np.delete(data, missing) x_missing = np.reshape(np.delete(x_pos, missing),-1) y_missing = np.reshape(np.delete(y_pos, missing),-1) data_map = get_data_map(data_missing, x_missing, y_missing) plot_spatial_map(data_map, x_missing, y_missing, alpha_map=data_map)

- aopy.visualization.base.plot_targets(target_positions, target_radius, bounds=None, alpha=0.5, origin=(0, 0, 0), ax=None, unique_only=True)[source]
Add targets to an axis. If any targets are at the origin, they will appear in a different color (magenta). Works for 2D and 3D axes
Example
Plot four peripheral and one central target.
target_position = np.array([ [0, 0, 0], [1, 1, 0], [-1, 1, 0], [1, -1, 0], [-1, -1, 0] ]) target_radius = 0.1 plot_targets(target_position, target_radius, (-2, 2, -2, 2))

- Parameters:
target_positions (ntarg, 3) – array of target (x, y, z) locations
target_radius (float) – radius of each target
bounds (tuple, optional) – 6-element tuple describing (-x, x, -y, y, -z, z) cursor bounds
origin (tuple, optional) – (x, y, z) position of the origin
ax (plt.Axis, optional) – axis to plot the targets on
unique_only (bool, optional) – If True, function will only plot targets with unique positions (default: True)
- aopy.visualization.base.plot_tfr(values, times, freqs, cmap='plasma', logscale=False, ax=None, **kwargs)[source]
Plot a time-frequency representation of a signal.
- Parameters:
values ((nt, nfreq) array) –
times ((nt,) array) –
freqs ((nfreq,) array) –
cmap (str, optional) – colormap to use for plotting
logscale (bool, optional) – apply a log scale to the color axis. Default False.
ax (pyplot.Axes, optional) – axes on which to plot. Default current axis.
kwargs (dict, optional) – other keyword arguments to pass to pyplot
- Returns:
image object returned from pyplot.pcolormesh. Useful for adding colorbars, etc.
- Return type:
pyplot.Image
Examples
fig, ax = plt.subplots(3,1,figsize=(4,6)) samplerate = 1000 data_200_hz = aopy.utils.generate_multichannel_test_signal(2, samplerate, 8, 200, 2) nt = data_200_hz.shape[0] data_200_hz[:int(nt/3),:] /= 3 data_200_hz[int(2*nt/3):,:] *= 2 data_50_hz = aopy.utils.generate_multichannel_test_signal(2, samplerate, 8, 50, 2) data_50_hz[:int(nt/2),:] /= 2 data = data_50_hz + data_200_hz print(data.shape) aopy.visualization.plot_timeseries(data, samplerate, ax=ax[0]) aopy.visualization.plot_freq_domain_amplitude(data, samplerate, ax=ax[1]) freqs = np.linspace(1,250,100) coef = aopy.analysis.calc_cwt_tfr(data, freqs, samplerate, fb=10, f0_norm=1, verbose=True) t = np.arange(nt)/samplerate print(data.shape) print(coef.shape) print(t.shape) print(freqs.shape) pcm = aopy.visualization.plot_tfr(abs(coef[:,:,0]), t, freqs, 'plasma', ax=ax[2]) fig.colorbar(pcm, label='Power', orientation = 'horizontal', ax=ax[2])

See also
calc_cwt_tfr()
- aopy.visualization.base.plot_timeseries(data, samplerate, t0=0.0, ax=None, **kwargs)[source]
Plots data along time on the given axis. Default units are seconds and volts.
Example
Plot 50 and 100 Hz sine wave.
data = np.reshape(np.sin(np.pi*np.arange(1000)/10) + np.sin(2*np.pi*np.arange(1000)/10), (1000)) samplerate = 1000 plot_timeseries(data, samplerate)

- Parameters:
data (nt, nch) – timeseries data in volts, can also be a single channel vector
samplerate (float) – sampling rate of the data
t0 (float, optional) – time (in seconds) of the first sample. Default 0.
ax (pyplot axis, optional) – where to plot
kwargs (dict, optional) – optional keyword arguments to pass to plt.plot
- aopy.visualization.base.plot_trajectories(trajectories, bounds=None, ax=None, **kwargs)[source]
Draws the given trajectories, one at a time in different colors. Works for 2D and 3D axes
Example
Two random trajectories.
trajectories =[ np.array([ [0, 0, 0], [1, 1, 0], [2, 2, 0], [3, 3, 0], [4, 2, 0] ]), np.array([ [-1, 1, 0], [-2, 2, 0], [-3, 3, 0], [-3, 4, 0] ]) ] bounds = (-5., 5., -5., 5., 0., 0.) plot_trajectories(trajectories, bounds)

- Parameters:
trajectories (list) – list of (n, 2) or (n, 3) trajectories where n can vary across each trajectory
bounds (tuple, optional) – 6-element tuple describing (-x, x, -y, y, -z, z) cursor bounds
ax (plt.Axis, optional) – axis to plot the targets on
kwargs (dict) – keyword arguments to pass to the plt.plot function
- aopy.visualization.base.plot_tuning_curves(fit_params, mean_fr, targets, n_subplot_cols=5, ax=None)[source]
This function plots the tuning curves output from analysis.run_tuningcurve_fit overlaying the actual firing rate data. The dashed line is the model fit and the solid line is the actual data.

- Parameters:
fit_params (nunits, 3) – Model fit coefficients. Output from analysis.run_tuningcurve_fit or analysis.curve_fitting_func
mean_fr (nunits, ntargets) – The average firing rate for each unit for each target.
target_theta (ntargets) – Orientation of each target in a center out task [degrees]. Corresponds to order of targets in ‘mean_fr’
n_subplot_cols (int) – Number of columns to plot in subplot. This function will automatically calculate the number of rows. Defaults to 5
ax (axes handle) – Axes to plot
- aopy.visualization.base.plot_waveforms(waveforms, samplerate, plot_mean=True, ax=None)[source]
This function plots the input waveforms on the same figure and can overlay the mean if requested

- Parameters:
waveforms (nt, nwfs) – Array of waveforms to plot
samplerate (float) – Sampling rate of waveforms to calculate time axis. [Hz]
plot_mean (bool) – Indicate if the mean waveform should be plotted. Defaults to plot mean.
ax (axes handle) – Axes to plot
- aopy.visualization.base.profile_data_channels(data, samplerate, figuredir, **kwargs)[source]
Runs plot_channel_summary and combine_channel_figures on all channels in a data array
- Parameters:
data (nt, nch) – numpy array of neural data
samplerate (int) – sampling rate of data
figuredir (str) – string indicating file path to desired save directory
kwargs (**dict) – keyword arguments to pass to plot_channel_summary()

- aopy.visualization.base.reset_plot_color(ax)[source]
Utility to reset the color cycle on a given axis to the default.
- Parameters:
ax (pyplot.Axes) – specify which axis to reset the color
Examples
Using reset_plot_color to reset the color cycle between calls to plt.plot().
plt.subplots() plt.plot(np.arange(10), np.ones(10)) aopy.visualization.reset_plot_color(plt.gca()) plt.plot(np.arange(10), 1 + np.ones(10))

- aopy.visualization.base.savefig(base_dir, filename, **kwargs)[source]
Wrapper around matplotlib savefig with some default options
- Parameters:
base_dir (str) – where to put the figure
filename (str) – what to name the figure
**kwargs (optional) – arguments to pass to plt.savefig()
- aopy.visualization.base.set_bounds(bounds, ax=None)[source]
Sets the x, y, and z limits according to the given bounds
- Parameters:
bounds (tuple) – 6-element tuple describing (-x, x, -y, y, -z, z) cursor bounds
ax (plt.Axis, optional) – axis to plot the targets on
- aopy.visualization.base.subplots_with_labels(n_rows, n_cols, return_labeled_axes=False, rel_label_x=-0.25, rel_label_y=1.1, label_font_size=11, constrained_layout=False, **kwargs)[source]
Create a figure with subplots labeled with letters. Augments plt.subplots().
Examples
Generate a figure with 2 rows and 2 columns of subplots, labeled A, B, C, D
fig, axes = subplots_with_labels(2, 2, constrained_layout=True)

- Parameters:
n_rows (int) – Number of rows of subplots.
n_cols (int) – Number of columns of subplots.
return_labeled_axes (bool, optional) – Whether to return the labeled axes. Default False.
rel_label_x (float, optional) – The relative x position of the subplot label. Default -0.25.
rel_label_y (float, optional) – The relative y position of the subplot label. Default 1.1
label_font_size (int, optional) – The font size of the subplot label. Default 11.
constrained_layout (bool, optional) – Whether to use constrained layout. Default is False.
**kwargs – Additional keyword arguments to pass to plt.subplot_mosaic.
- Returns:
The created figure. axes (np.ndarray): The created axes. labels_axes (dict, optional): The labeled axes if return_labeled_axes is True.
- Return type:
fig (Figure)
Animation
- aopy.visualization.animation.animate_behavior(targets, cursor, eye, samplerate, bounds, target_radius, target_colors, cursor_radius, cursor_color='blue', eye_radius=0.25, eye_color='purple', history=0.0)[source]
Animate target, cursor, and eye data together.
- Parameters:
targets (list of (nt,) arrays) – Target position timeseires for each target.
cursor ((nt, 2) array) – Cursor position timeseires.
eye ((nt, 2) array) – Eye position timeseires.
samplerate (float) – The sampling rate of all the trajectories in Hz.
bounds (tuple) – Boundaries of the plot area. See
plot_targets().target_radius (float) – Radius of the targets.
target_colors (list of plt.color) – Color of each target.
cursor_radius (float) – Radius of the cursor.
cursor_color (plt.color, optional) – Color of the cursor. Default is ‘blue’.
eye_radius (float) – Radius of the eye circle.
eye_color (plt.color, optional) – Color of the eye trajectory. Default is ‘purple’.
history (float, optional) – how long (in seconds) to animate lines trailing the circles. Default 0.
- Returns:
animation object
- Return type:
matplotlib.animation.FuncAnimation
Example
samplerate = 0.5 cursor = np.array([[0,0], [1, 2], [2, 3], [3, 4], [4, 5], [5, 6]]) eye = np.array([[1, 0], [1, 2], [1, 2], [4, 5], [4, 5], [6, 6]]) targets = [ np.array([[np.nan, np.nan], [5, 5], [np.nan, np.nan], [np.nan, np.nan], [5, 5], [np.nan, np.nan]]), np.array([[np.nan, np.nan], [np.nan, np.nan], [np.nan, np.nan], [-5, 5], [-5, 5], [-5, 5]]) ] target_radius = 2.5 target_colors = ['orange'] * len(targets) cursor_radius = 0.5 bounds = [-10, 10, -10, 10] ani = animate_behavior(targets, cursor, eye, samplerate, bounds, target_radius, target_colors, cursor_radius, cursor_color='blue', eye_radius=0.25, eye_color='purple')
- aopy.visualization.animation.animate_cursor_eye(cursor_trajectory, eye_trajectory, samplerate, target_positions, target_radius, bounds, cursor_radius=0.5, eye_radius=0.25, cursor_color='blue', eye_color='purple')[source]
Draws an animation of two trajectories with static targets. The colors and endpoint radii of the two trajectories can be specified along with the position and radius of the targets. Targets are colored automatically according to
plot_targets().Example
- Parameters:
cursor_trajectory ((nt, ndim) array) – Cursor positions over time for 2D or 3D trajectories.
eye_trajectory ((nt, ndim) array) – Eye positions over time for 2D or 3D trajectories.
samplerate (float) – The sampling rate of the trajectories in Hz.
target_positions ((ntargets, ndim) array) – Array of target positions for 2D or 3D targets.
target_radius (float) – Radius of the targets.
bounds (tuple) – Boundaries of the plot area. See
plot_targets().cursor_radius (float, optional) – Radius of the cursor endpoint. Default is 0.5.
eye_radius (float, optional) – Radius of the eye endpoint. Default is 0.25.
cursor_color (plt.color, optional) – Color of the cursor trajectory. Default is ‘blue’.
eye_color (plt.color, optional) – Color of the eye trajectory. Default is ‘purple’.
- Returns:
None
- Returns:
animation object
- Return type:
matplotlib.animation.FuncAnimation
- aopy.visualization.animation.animate_events(events, times, fps, xy=(0.3, 0.3), fontsize=30, color='g')[source]
Silly function to plot events as text, frame by frame in an animation
- Parameters:
events (list) – list of event names or numbers
times (list) – timestamps of each event
fps (float) – sampling rate to animate
xy (tuple, optional) – (x, y) coorindates of the left bottom corner of each event label, from 0 to 1.
fontsize (float, optional) – size to draw the event labels
- Returns:
animation object
- Return type:
matplotlib.animation.FuncAnimation
Example
- aopy.visualization.animation.animate_spatial_map(data_map, x, y, samplerate, cmap='bwr')[source]
Animates a 2d heatmap. Use
aopy.visualization.get_data_map()to get a 2d array for each timepoint you want to animate, then put them into a list and feed them to this function. See alsoaopy.visualization.show_anim()andaopy.visualization.save_anim()Example
samplerate = 20 duration = 5 x_pos, y_pos = np.meshgrid(np.arange(0.5,10.5),np.arange(0.5, 10.5)) data_map = [] for frame in range(duration*samplerate): t = np.linspace(-1, 1, 100) + float(frame)/samplerate c = np.sin(t) data_map.append(get_data_map(c, x_pos.reshape(-1), y_pos.reshape(-1))) filename = 'spatial_map_animation.mp4' ani = animate_spatial_map(data_map, x_pos, y_pos, samplerate, cmap='bwr') saveanim(ani, write_dir, filename)
- Parameters:
data_map (nt) – array of 2d maps
x (list) – list of x positions
y (list) – list of y positions
samplerate (float) – rate of the data_map samples
cmap (str, optional) – name of the colormap to use. Defaults to ‘bwr’.
- Returns:
animation object
- Return type:
matplotlib.animation.FuncAnimation
- aopy.visualization.animation.animate_trajectory_3d(trajectory, samplerate, history=1000, color='b', axis_labels=['x', 'y', 'z'])[source]
Draws a trajectory moving through 3D space at the given sampling rate and with a fixed maximum number of points visible at a time.
- Parameters:
trajectory (n, 3) – matrix of n points
samplerate (float) – sampling rate of the trajectory data
history (int, optional) – maximum number of points visible at once
- Returns:
animation object
- Return type:
matplotlib.animation.FuncAnimation
Example
- aopy.visualization.animation.get_animate_circles_func(samplerate, bounds, circle_radii, circle_colors, *circle_ts, history=1.0, ax=None)[source]
Draws an animation of an arbitrary number of circles. Used in
animate_behavior().- Parameters:
samplerate (float) – The sampling rate of the trajectories in Hz.
bounds (tuple) – Boundaries of the plot area. See
plot_targets().circle_radii (list of float) – Radius of each circle.
circle_colors (list of plt.color) – Color of each circle.
circle_ts (list of (nt, 2) arrays) – Circle positions over time for 2D trajectories.
history (float, optional) – how long (in seconds) to animate lines trailing the circles. Default 1.
ax (pyplot.Axes, optional) – axis on which to plot the animation
- Returns:
plotting function for FuncAnimation
- Return type:
function
- aopy.visualization.animation.saveanim(animation, base_dir, filename, dpi=100, **savefig_kwargs)[source]
Save an animation using ffmpeg
- Parameters:
animation (pyplot.Animation) – animation to save
base_dir (str) – directory to write
filename (str) – should end in ‘.mp4’
dpi (float) – resolution of the video file
savefig_kwargs (kwargs, optional) – arguments to pass to savefig